Xenon, Xe »
PDB 4zzc-7tsj »
7sxm »
Xenon in PDB 7sxm: Structure of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis
Enzymatic activity of Structure of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis
All present enzymatic activity of Structure of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis:
2.8.4.1;
2.8.4.1;
Protein crystallography data
The structure of Structure of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis, PDB code: 7sxm
was solved by
P.Y.-T.Chen,
C.L.Drennan,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 69.63 / 2.50 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 81.956, 115.741, 123.4, 90, 92.53, 90 |
| R / Rfree (%) | 17.8 / 21.8 |
Other elements in 7sxm:
The structure of Structure of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis also contains other interesting chemical elements:
| Magnesium | (Mg) | 2 atoms |
| Nickel | (Ni) | 2 atoms |
| Sodium | (Na) | 1 atom |
Xenon Binding Sites:
The binding sites of Xenon atom in the Structure of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis
(pdb code 7sxm). This binding sites where shown within
5.0 Angstroms radius around Xenon atom.
In total 8 binding sites of Xenon where determined in the Structure of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis, PDB code: 7sxm:
Jump to Xenon binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
In total 8 binding sites of Xenon where determined in the Structure of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis, PDB code: 7sxm:
Jump to Xenon binding site number: 1; 2; 3; 4; 5; 6; 7; 8;
Xenon binding site 1 out of 8 in 7sxm
Go back to
Xenon binding site 1 out
of 8 in the Structure of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis
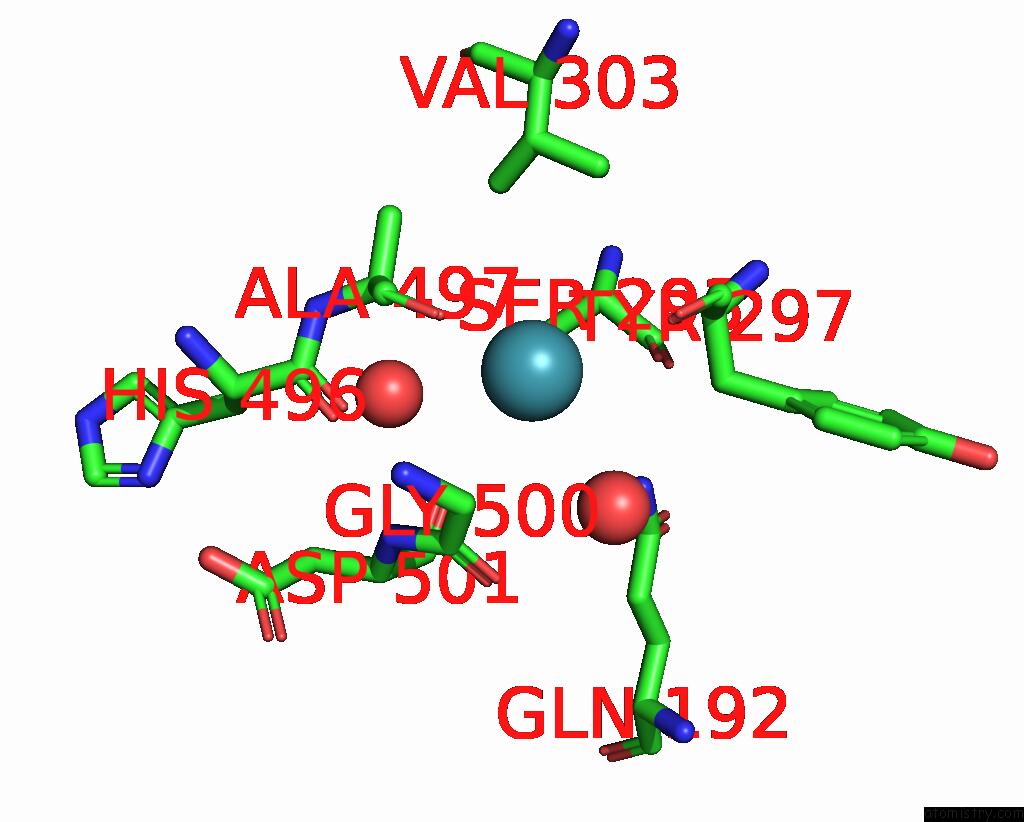
Mono view
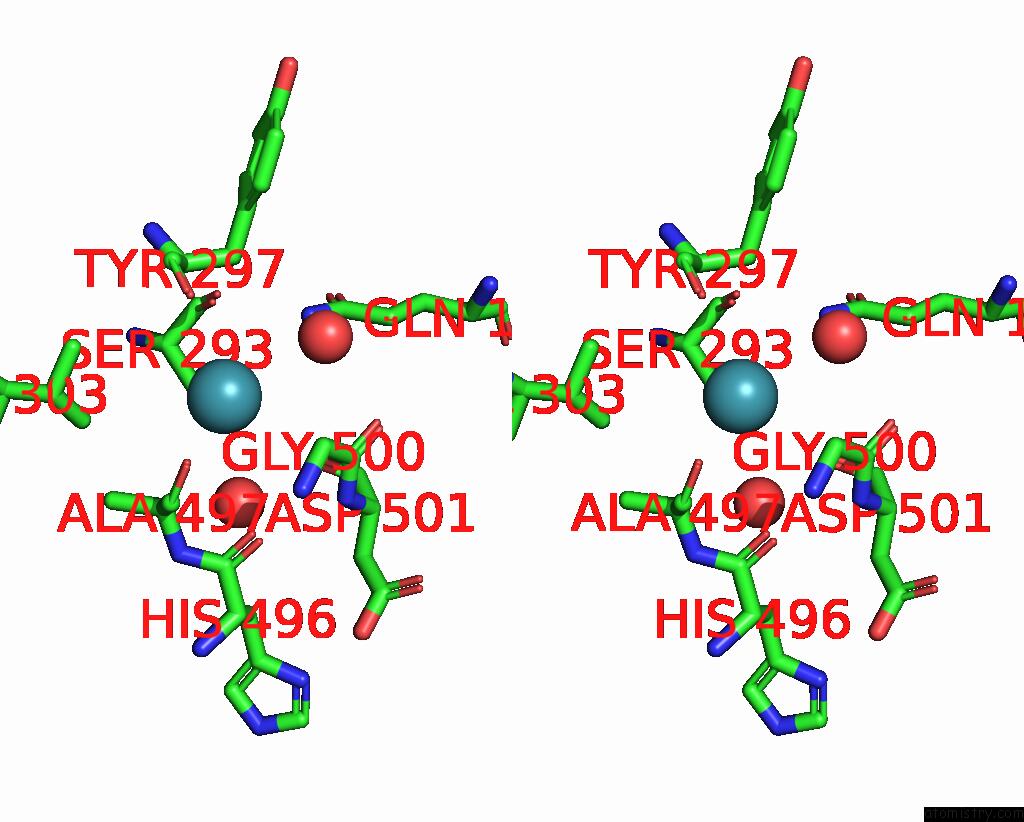
Stereo pair view

Mono view

Stereo pair view
A full contact list of Xenon with other atoms in the Xe binding
site number 1 of Structure of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis within 5.0Å range:
|
Xenon binding site 2 out of 8 in 7sxm
Go back to
Xenon binding site 2 out
of 8 in the Structure of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis
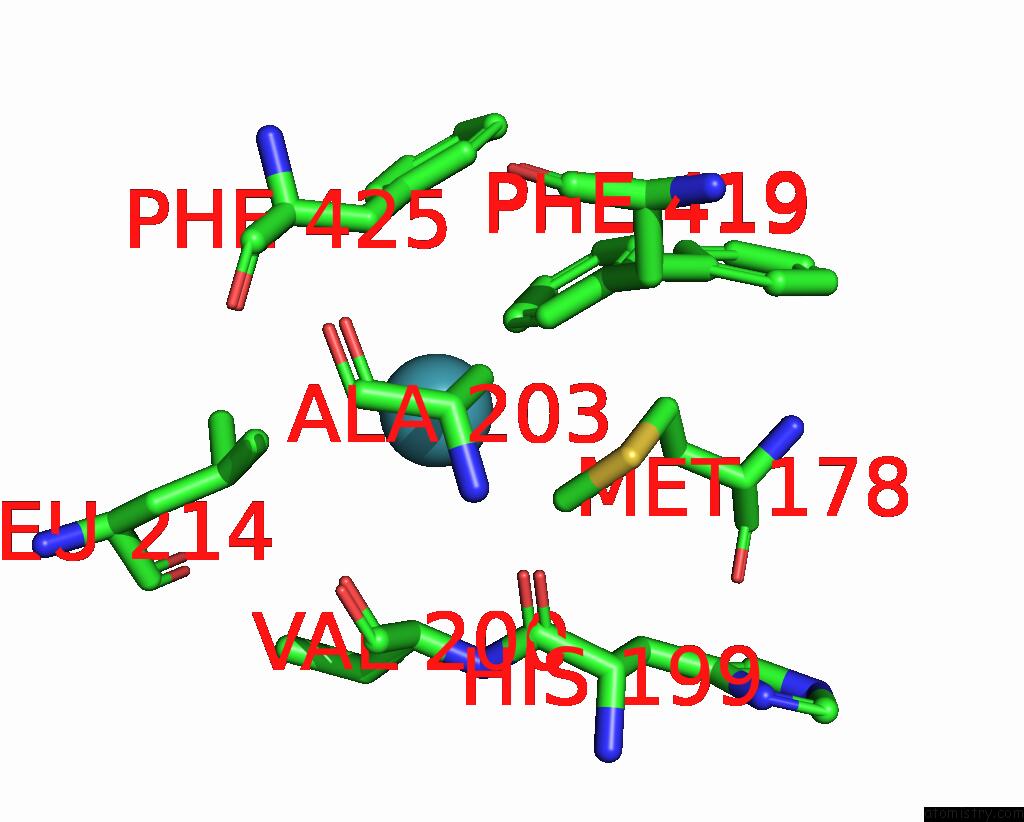
Mono view
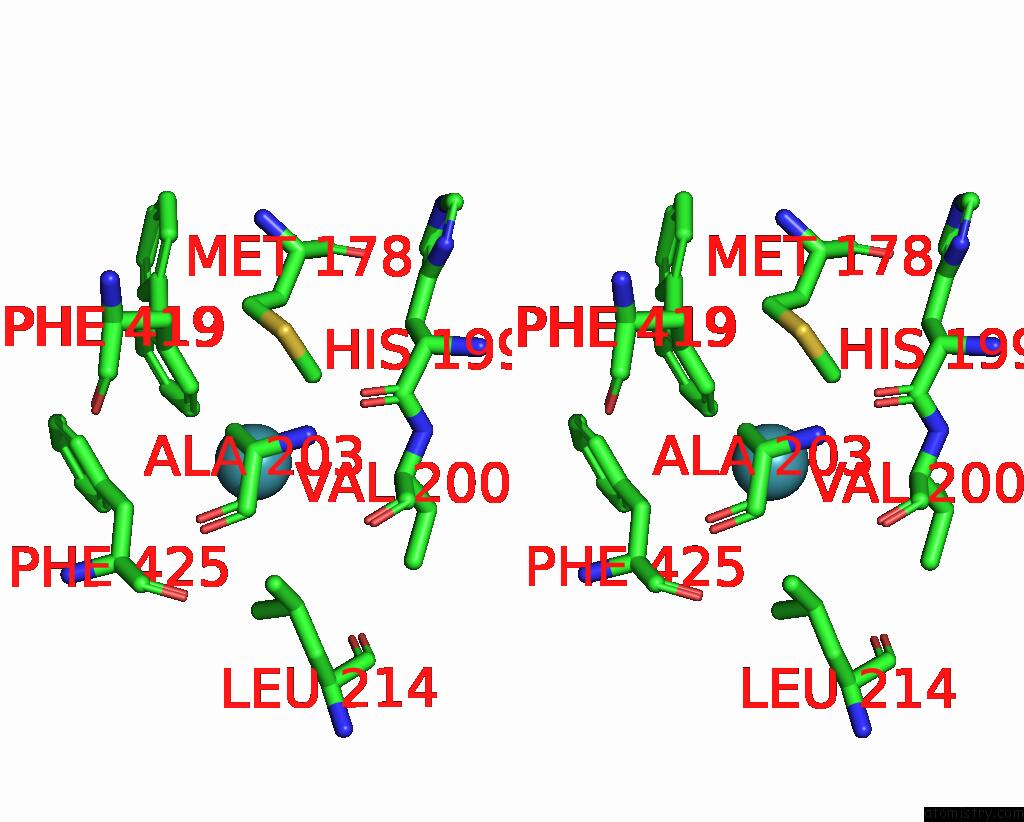
Stereo pair view

Mono view

Stereo pair view
A full contact list of Xenon with other atoms in the Xe binding
site number 2 of Structure of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis within 5.0Å range:
|
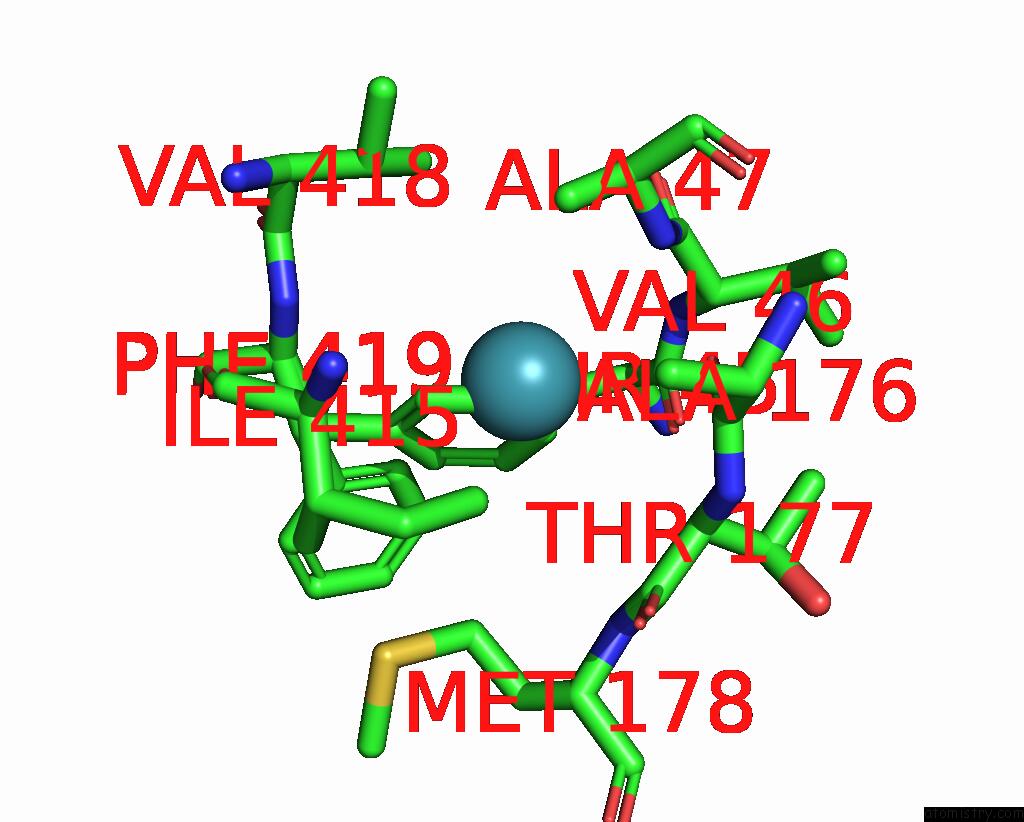
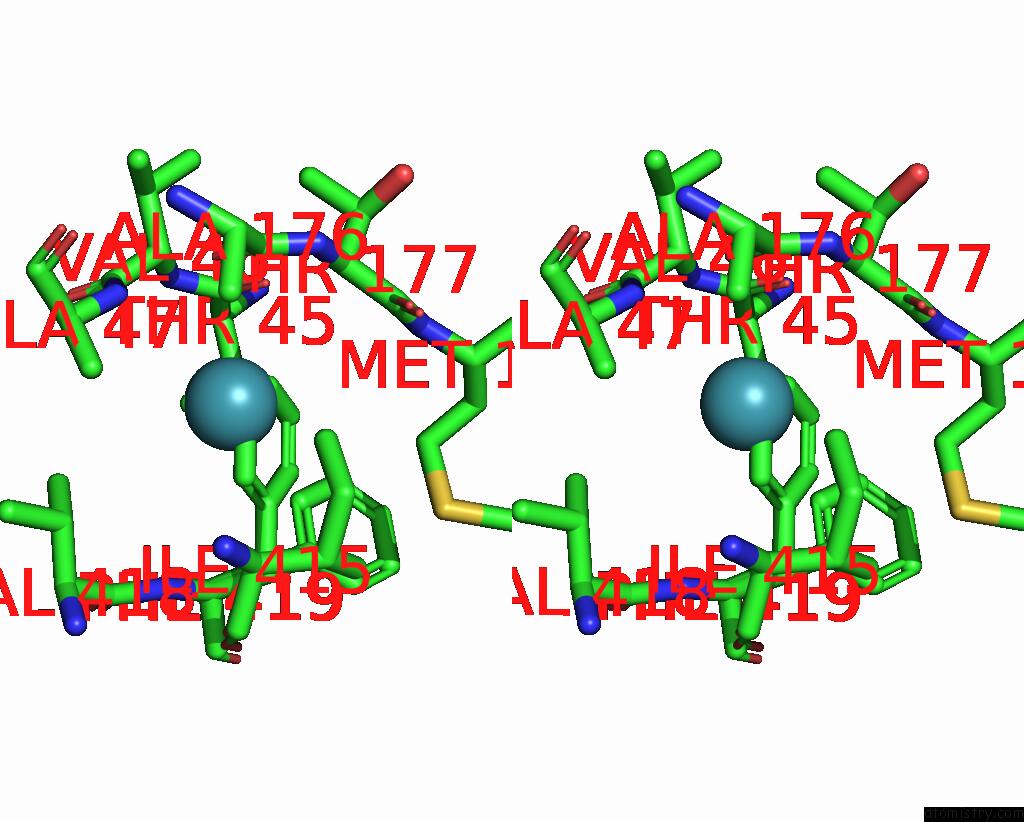
Xenon binding site 3 out of 8 in 7sxm
Go back to
Xenon binding site 3 out
of 8 in the Structure of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Xenon with other atoms in the Xe binding
site number 3 of Structure of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis within 5.0Å range:
|
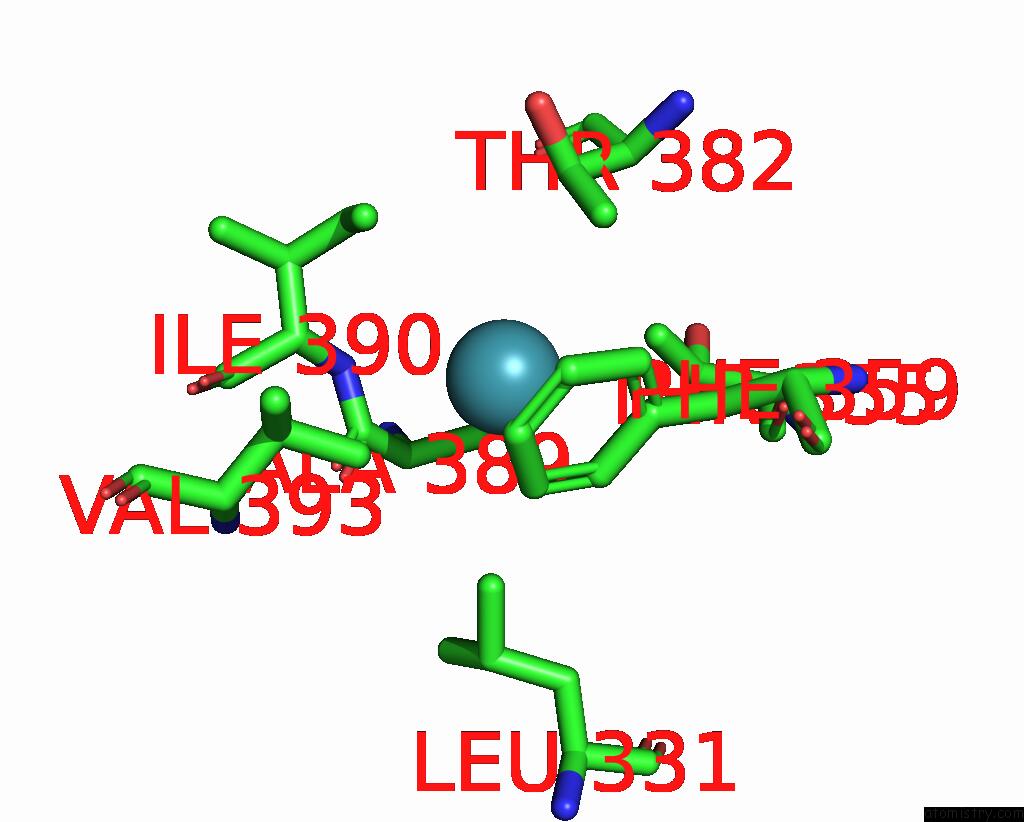
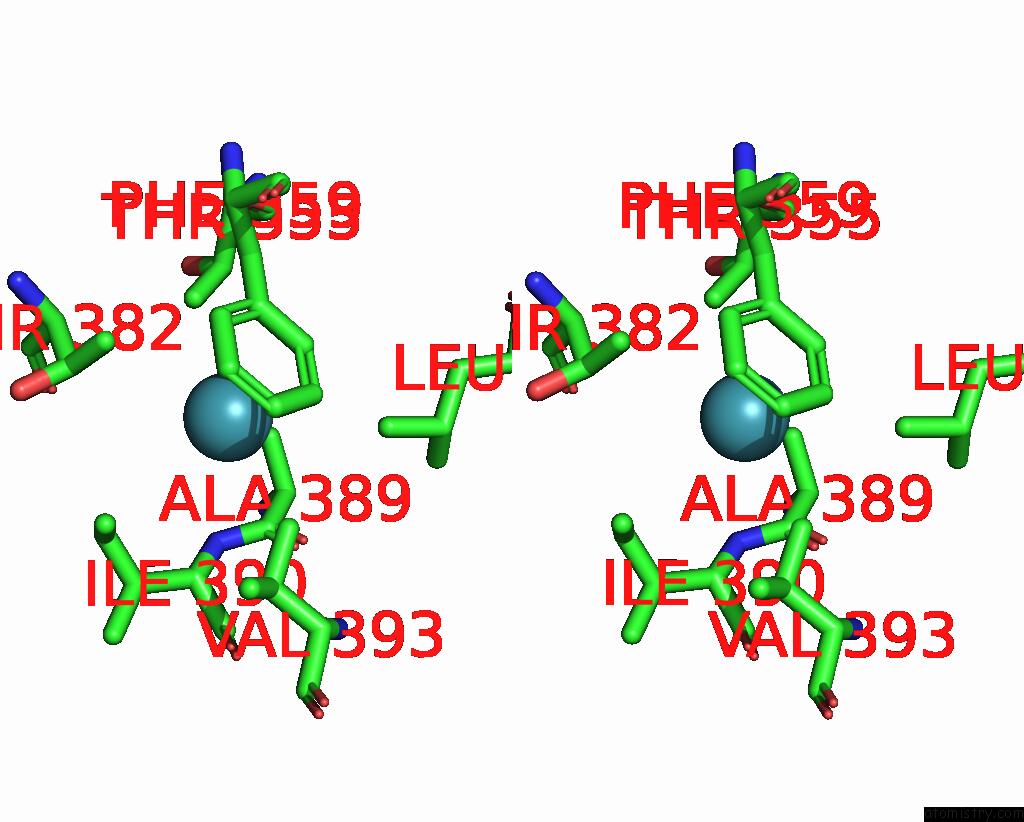
Xenon binding site 4 out of 8 in 7sxm
Go back to
Xenon binding site 4 out
of 8 in the Structure of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Xenon with other atoms in the Xe binding
site number 4 of Structure of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis within 5.0Å range:
|
Xenon binding site 5 out of 8 in 7sxm
Go back to
Xenon binding site 5 out
of 8 in the Structure of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis
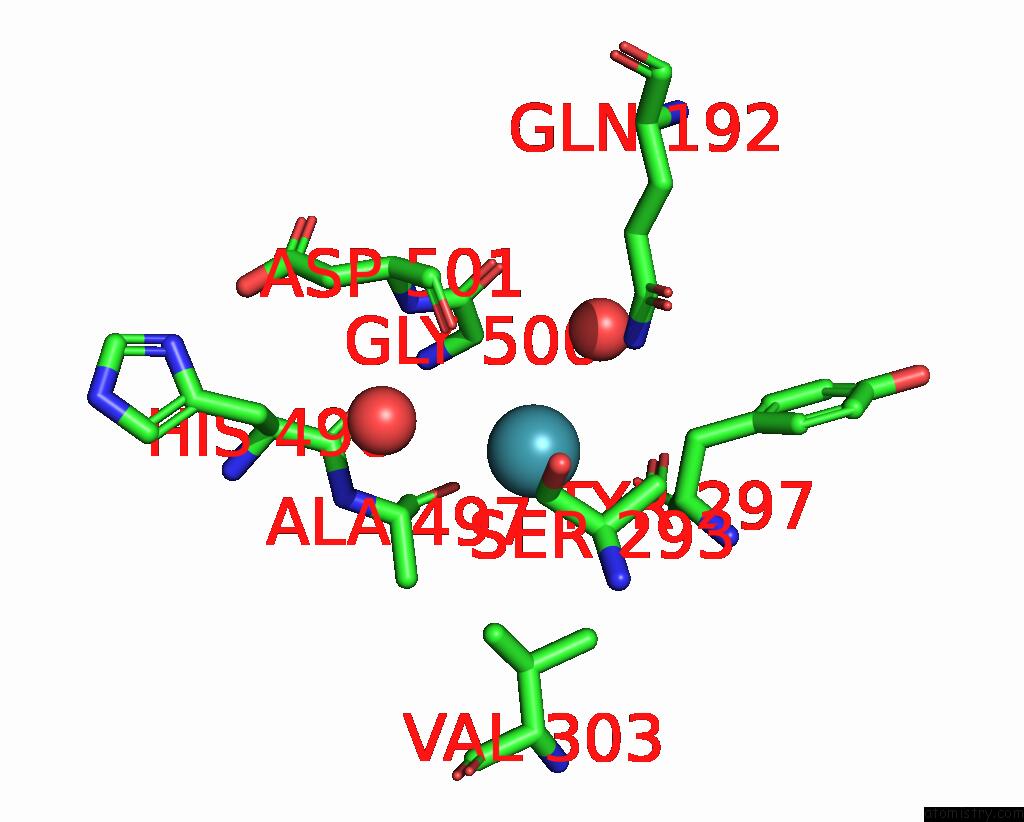
Mono view
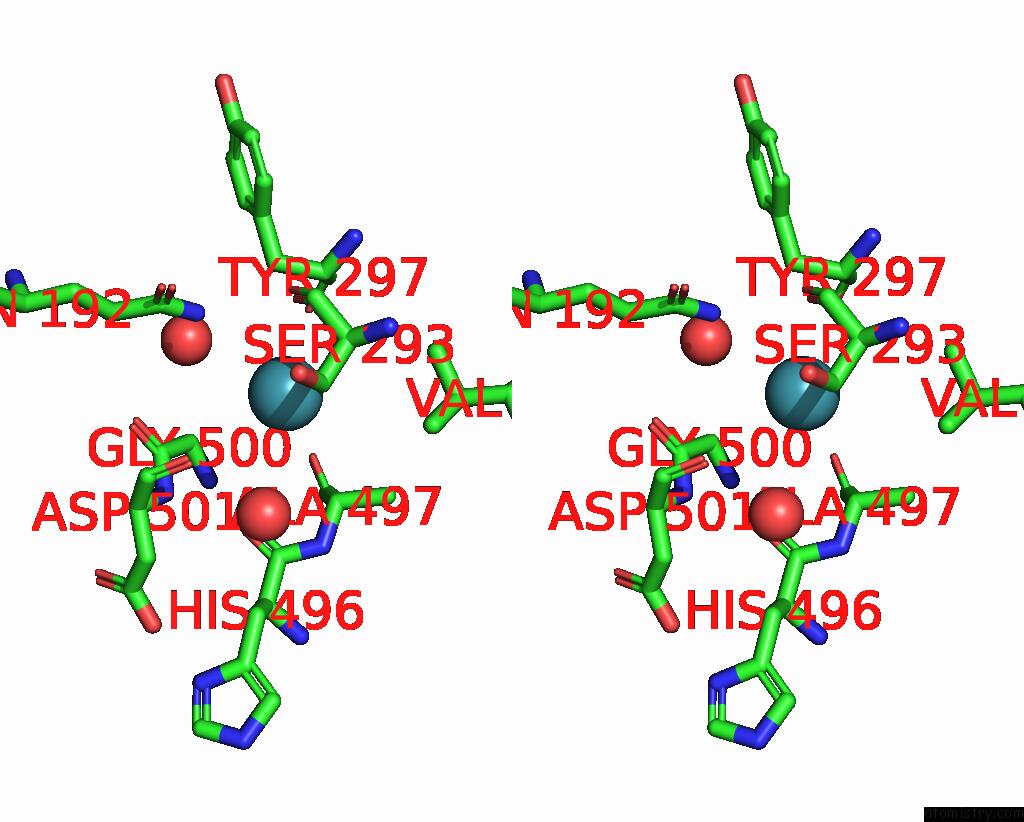
Stereo pair view

Mono view

Stereo pair view
A full contact list of Xenon with other atoms in the Xe binding
site number 5 of Structure of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis within 5.0Å range:
|
Xenon binding site 6 out of 8 in 7sxm
Go back to
Xenon binding site 6 out
of 8 in the Structure of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis
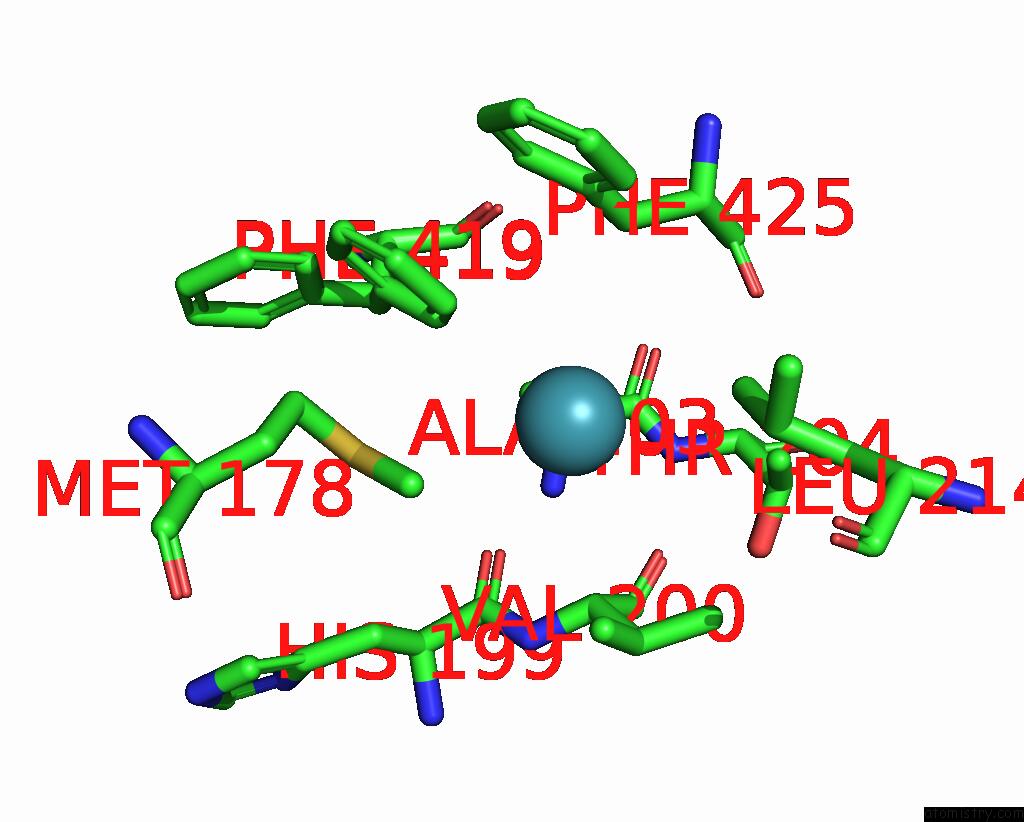
Mono view
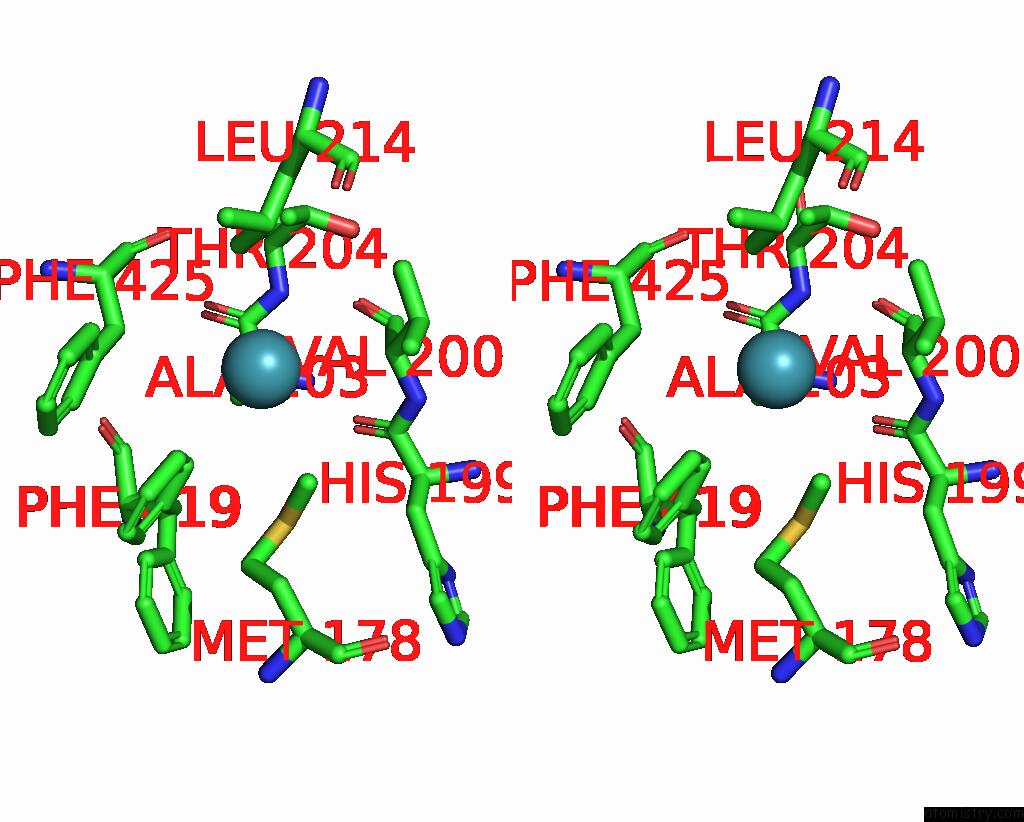
Stereo pair view

Mono view

Stereo pair view
A full contact list of Xenon with other atoms in the Xe binding
site number 6 of Structure of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis within 5.0Å range:
|
Xenon binding site 7 out of 8 in 7sxm
Go back to
Xenon binding site 7 out
of 8 in the Structure of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis
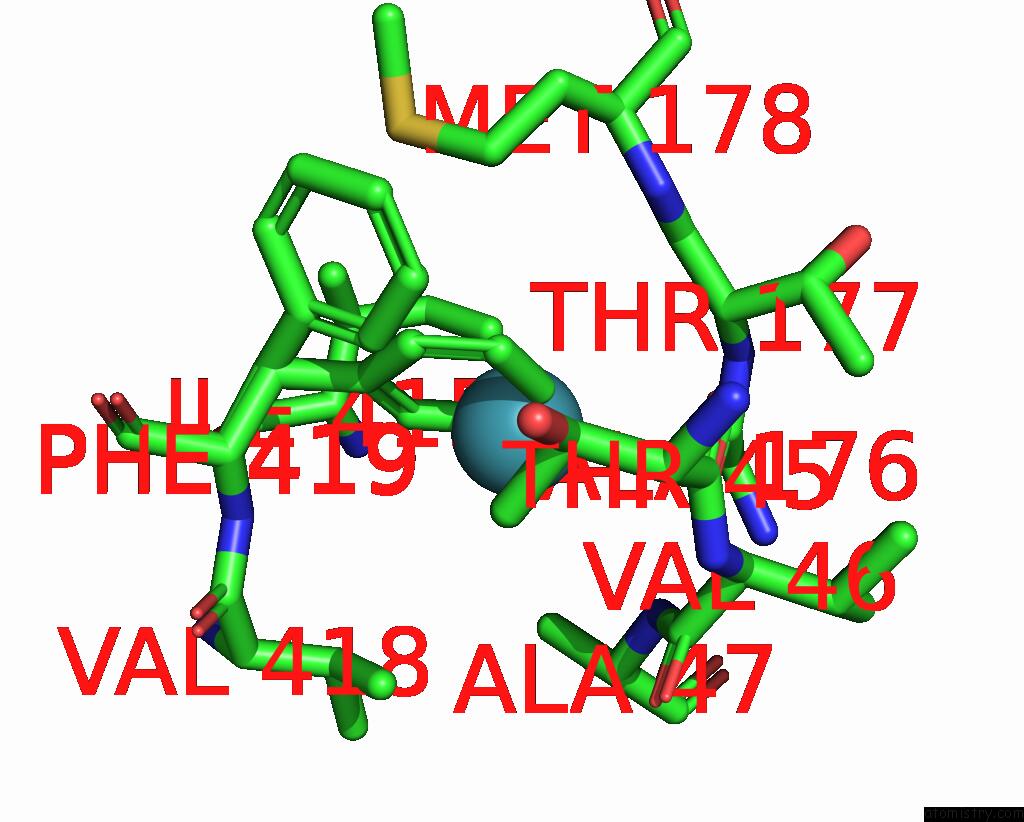
Mono view
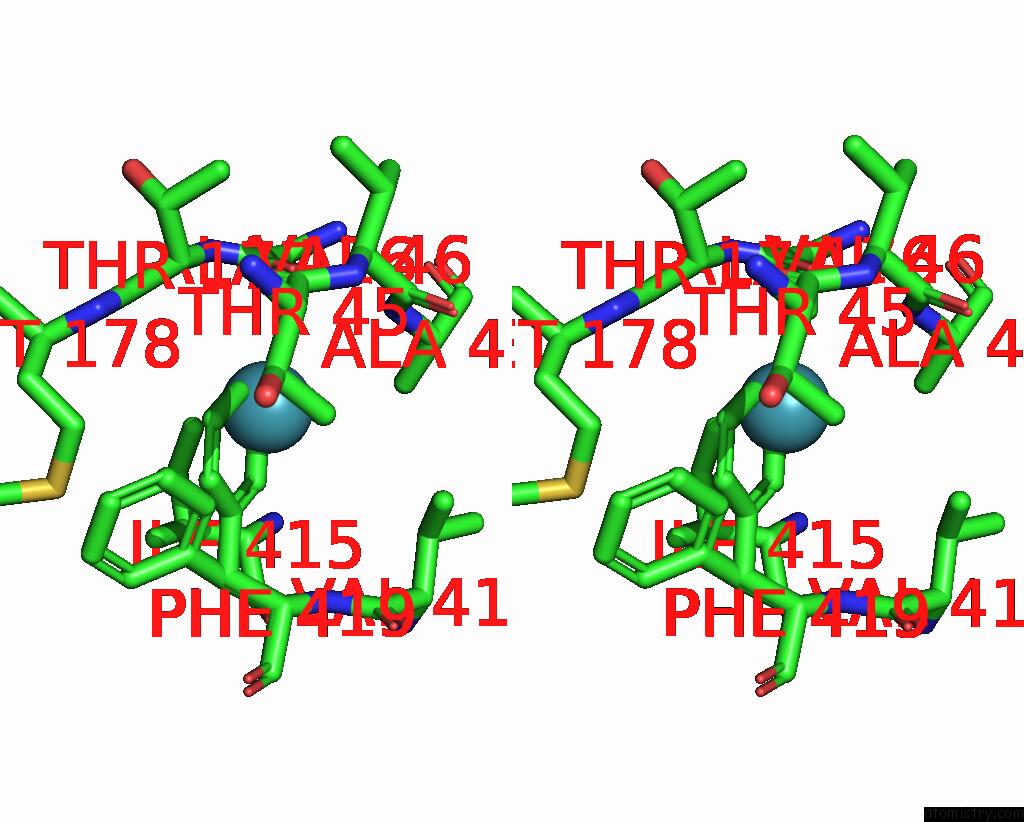
Stereo pair view

Mono view

Stereo pair view
A full contact list of Xenon with other atoms in the Xe binding
site number 7 of Structure of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis within 5.0Å range:
|
Xenon binding site 8 out of 8 in 7sxm
Go back to
Xenon binding site 8 out
of 8 in the Structure of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis
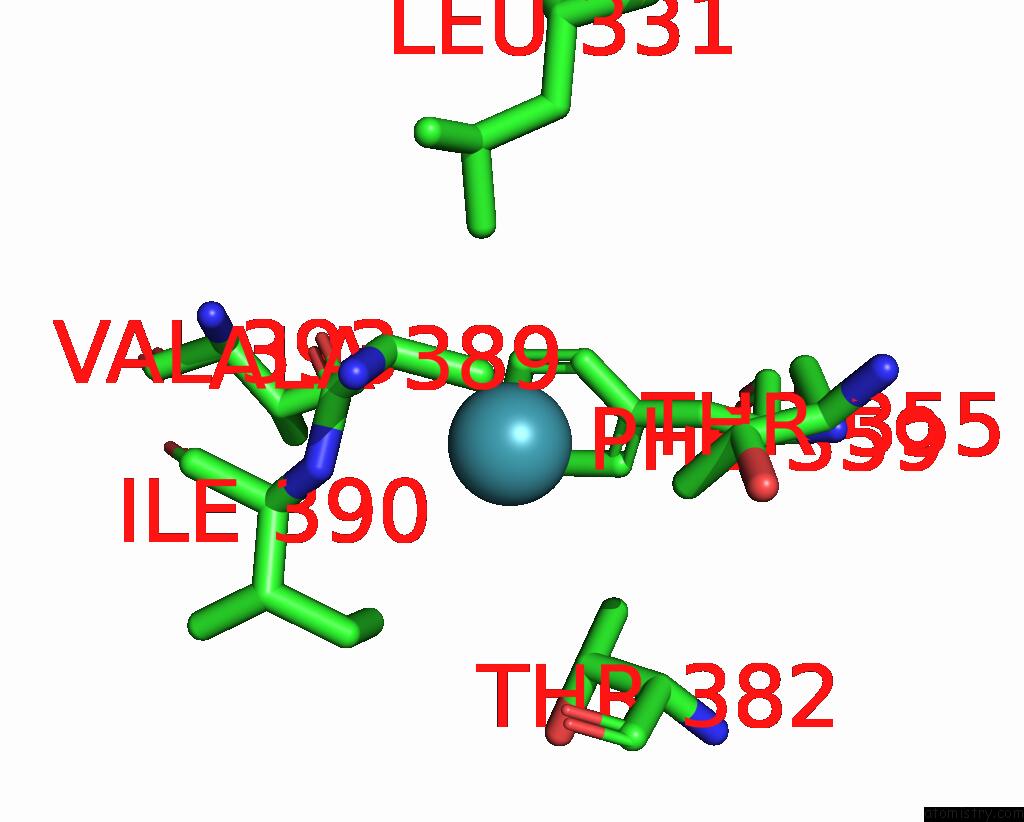
Mono view
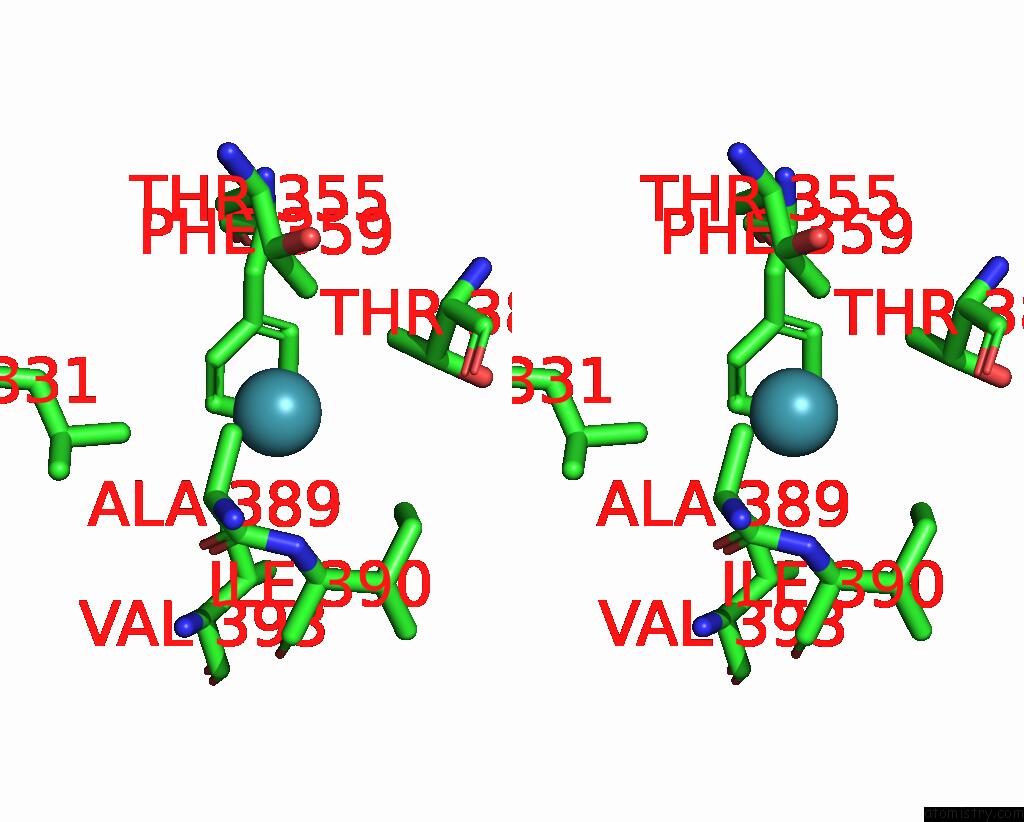
Stereo pair view

Mono view

Stereo pair view
A full contact list of Xenon with other atoms in the Xe binding
site number 8 of Structure of Xenon-Derivatized Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis within 5.0Å range:
|
Reference:
C.J.Ohmer,
M.Dasgupta,
A.Patwardhan,
I.Bogacz,
C.Kaminsky,
M.D.Doyle,
P.Y.Chen,
S.M.Keable,
H.Makita,
P.S.Simon,
R.Massad,
T.Fransson,
R.Chatterjee,
A.Bhowmick,
D.W.Paley,
N.W.Moriarty,
A.S.Brewster,
L.B.Gee,
R.Alonso-Mori,
F.Moss,
F.D.Fuller,
A.Batyuk,
N.K.Sauter,
U.Bergmann,
C.L.Drennan,
V.K.Yachandra,
J.Yano,
J.F.Kern,
S.W.Ragsdale.
Xfel Serial Crystallography Reveals the Room Temperature Structure of Methyl-Coenzyme M Reductase. J.Inorg.Biochem. V. 230 11768 2022.
ISSN: ISSN 0162-0134
PubMed: 35202981
DOI: 10.1016/J.JINORGBIO.2022.111768
Page generated: Sat Oct 12 20:05:52 2024
ISSN: ISSN 0162-0134
PubMed: 35202981
DOI: 10.1016/J.JINORGBIO.2022.111768
Last articles
F in 4MH0F in 4MDD
F in 4MDB
F in 4MC9
F in 4MDA
F in 4MC6
F in 4MC2
F in 4MC1
F in 4M8N
F in 4MBS