Xenon, Xe »
PDB 4zzc-9kum »
7shw »
Xenon in PDB 7shw: Crystal Structure of Mycobacterium Smegmatis Lmca with Xenon
Protein crystallography data
The structure of Crystal Structure of Mycobacterium Smegmatis Lmca with Xenon, PDB code: 7shw
was solved by
O.Patel,
I.Lucet,
S.Panjikar,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 48.58 / 1.79 |
| Space group | P 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 34.12, 56.989, 73.573, 78.42, 89.21, 78.92 |
| R / Rfree (%) | 19.7 / 21 |
Xenon Binding Sites:
The binding sites of Xenon atom in the Crystal Structure of Mycobacterium Smegmatis Lmca with Xenon
(pdb code 7shw). This binding sites where shown within
5.0 Angstroms radius around Xenon atom.
In total 5 binding sites of Xenon where determined in the Crystal Structure of Mycobacterium Smegmatis Lmca with Xenon, PDB code: 7shw:
Jump to Xenon binding site number: 1; 2; 3; 4; 5;
In total 5 binding sites of Xenon where determined in the Crystal Structure of Mycobacterium Smegmatis Lmca with Xenon, PDB code: 7shw:
Jump to Xenon binding site number: 1; 2; 3; 4; 5;
Xenon binding site 1 out of 5 in 7shw
Go back to
Xenon binding site 1 out
of 5 in the Crystal Structure of Mycobacterium Smegmatis Lmca with Xenon
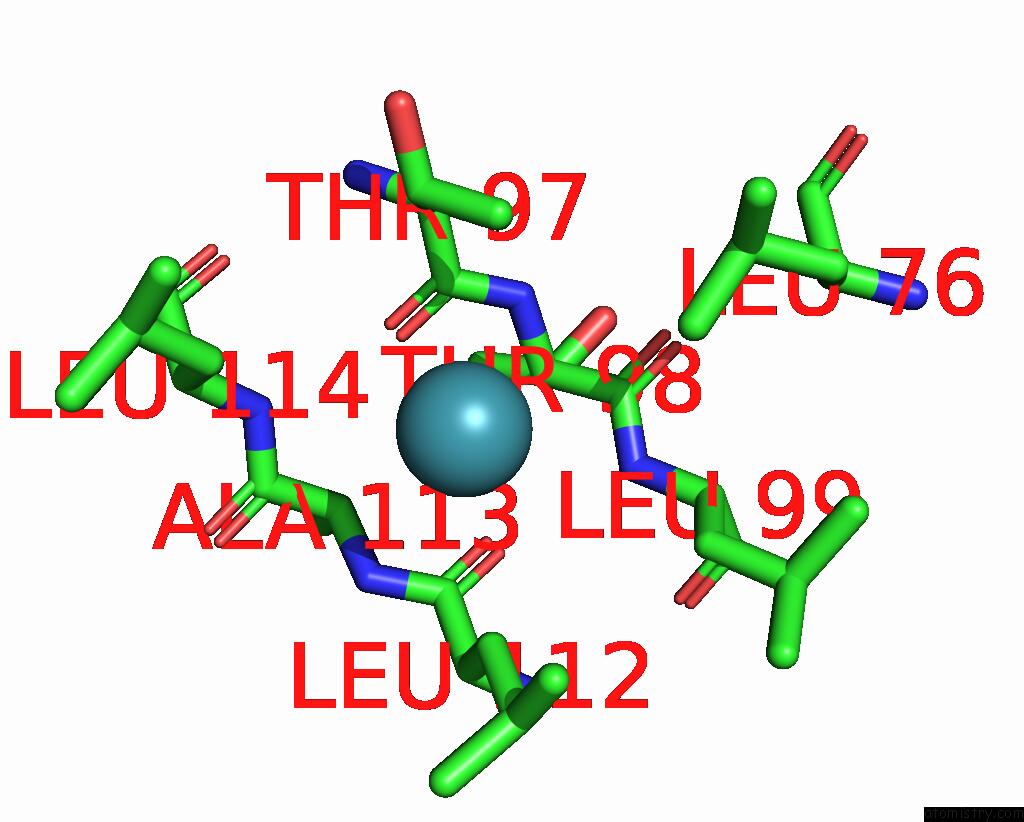
Mono view
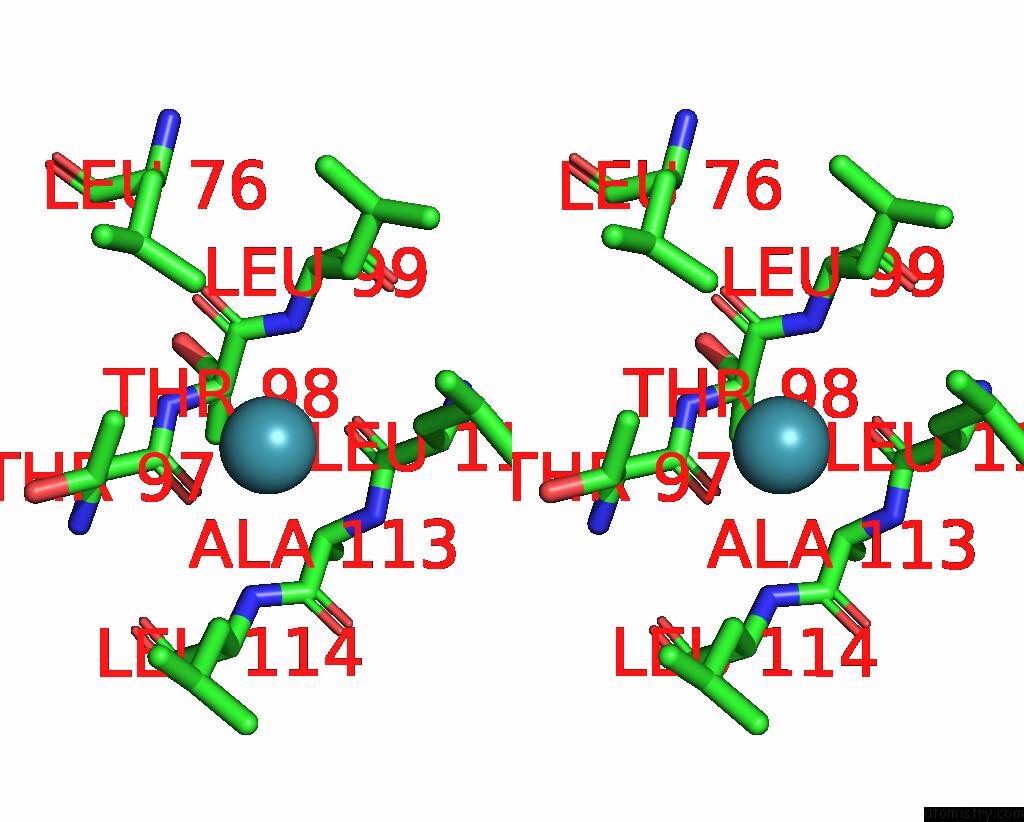
Stereo pair view

Mono view

Stereo pair view
A full contact list of Xenon with other atoms in the Xe binding
site number 1 of Crystal Structure of Mycobacterium Smegmatis Lmca with Xenon within 5.0Å range:
|
Xenon binding site 2 out of 5 in 7shw
Go back to
Xenon binding site 2 out
of 5 in the Crystal Structure of Mycobacterium Smegmatis Lmca with Xenon
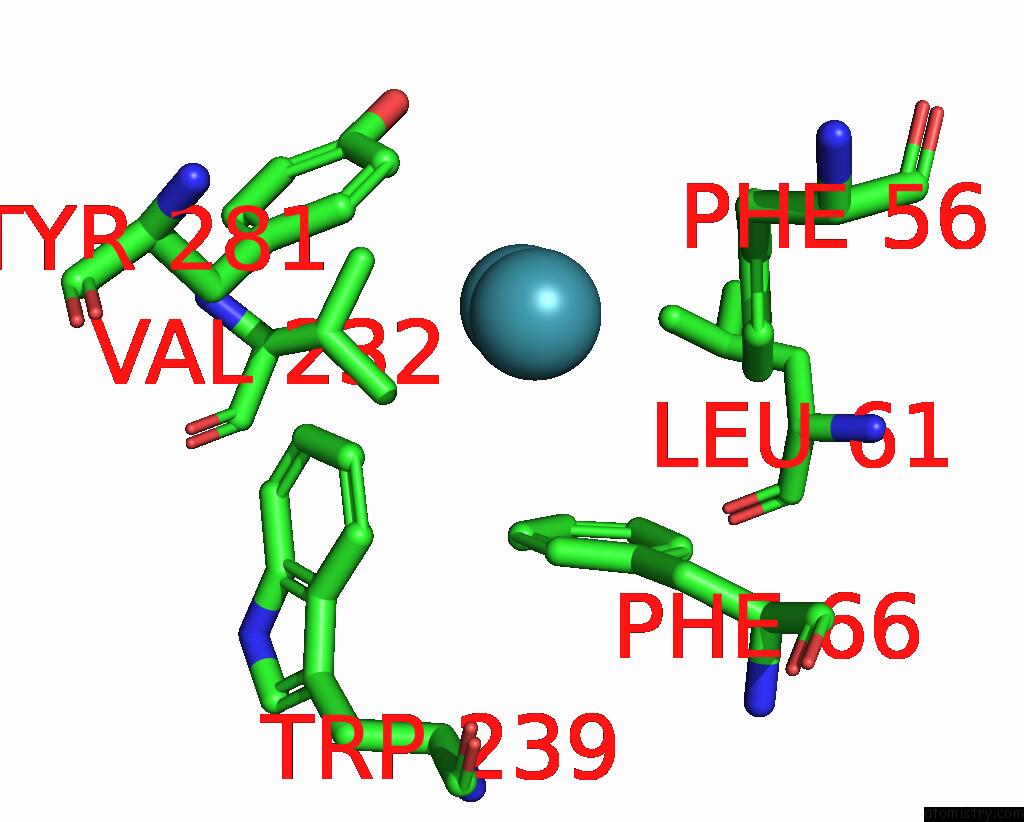
Mono view
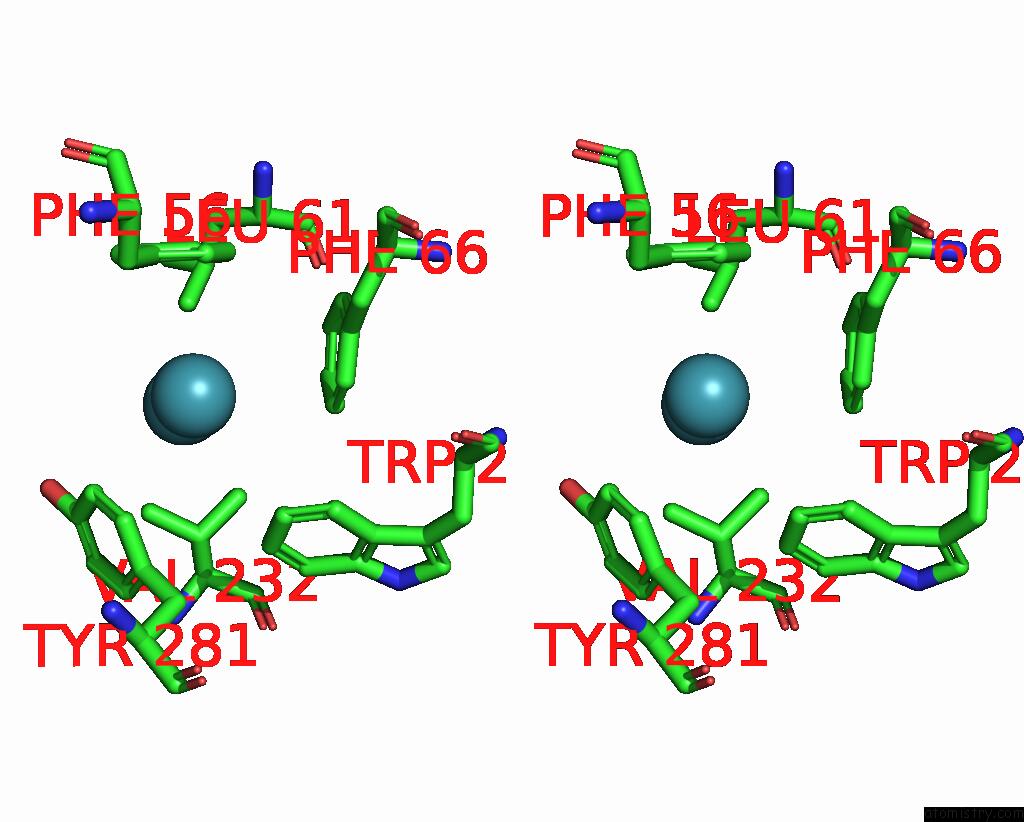
Stereo pair view

Mono view

Stereo pair view
A full contact list of Xenon with other atoms in the Xe binding
site number 2 of Crystal Structure of Mycobacterium Smegmatis Lmca with Xenon within 5.0Å range:
|
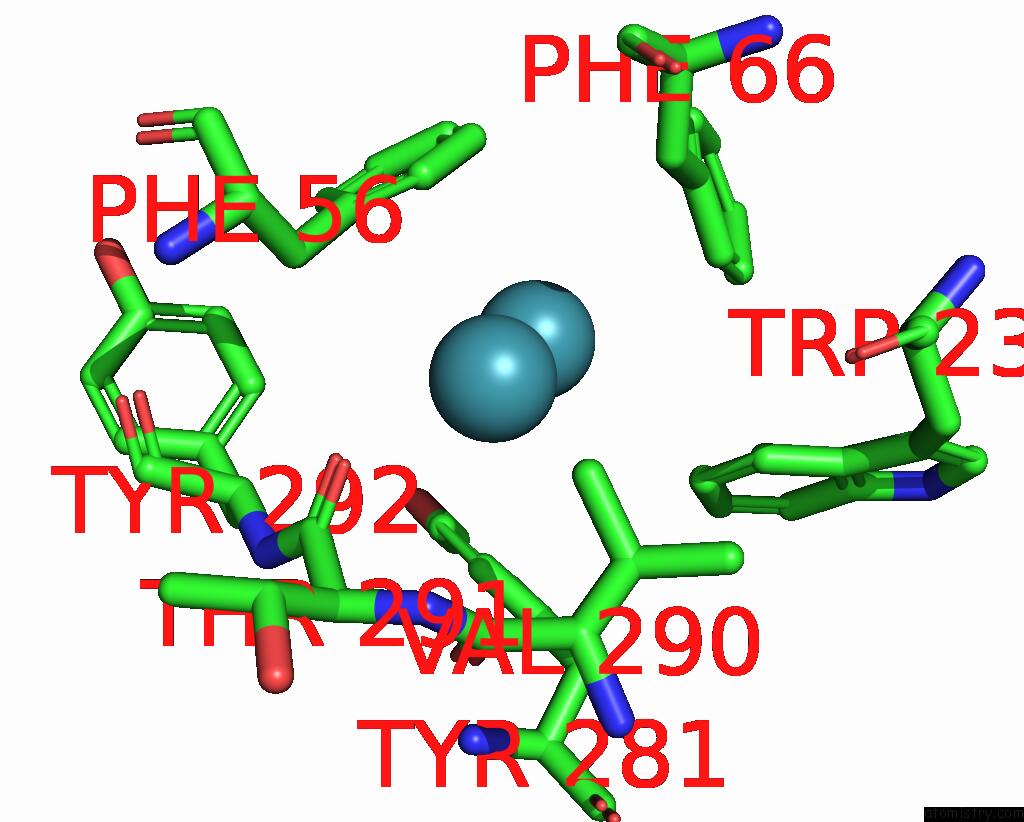
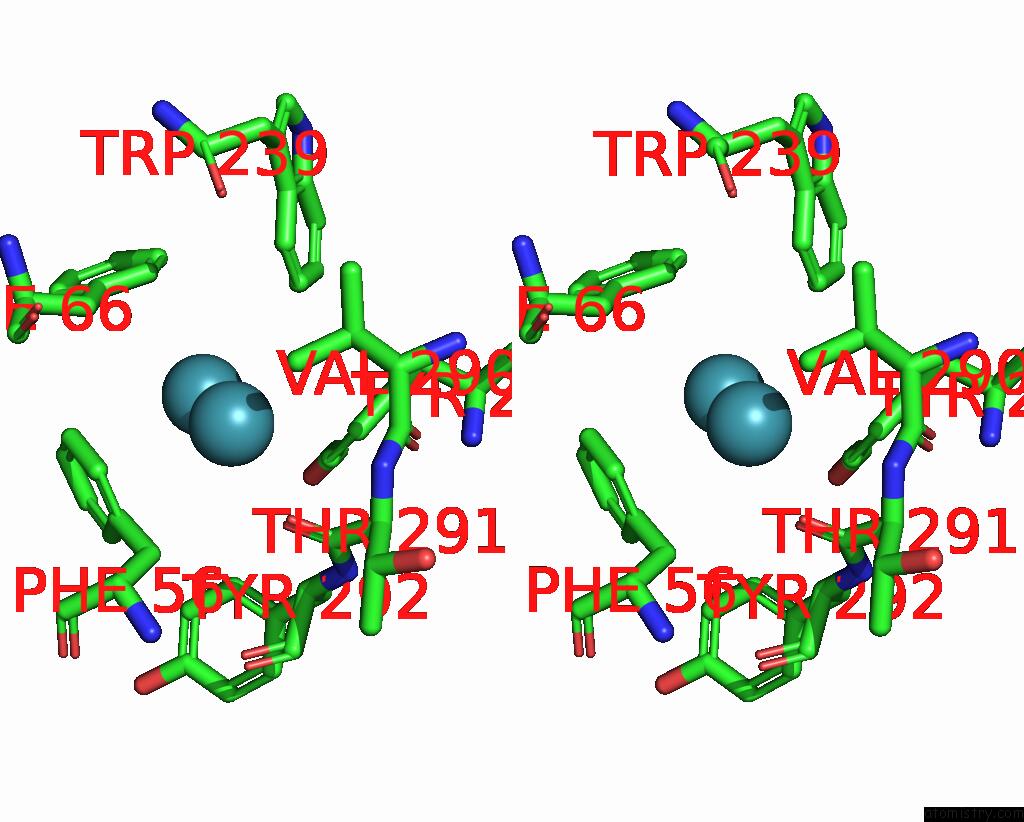
Xenon binding site 3 out of 5 in 7shw
Go back to
Xenon binding site 3 out
of 5 in the Crystal Structure of Mycobacterium Smegmatis Lmca with Xenon
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Xenon with other atoms in the Xe binding
site number 3 of Crystal Structure of Mycobacterium Smegmatis Lmca with Xenon within 5.0Å range:
|
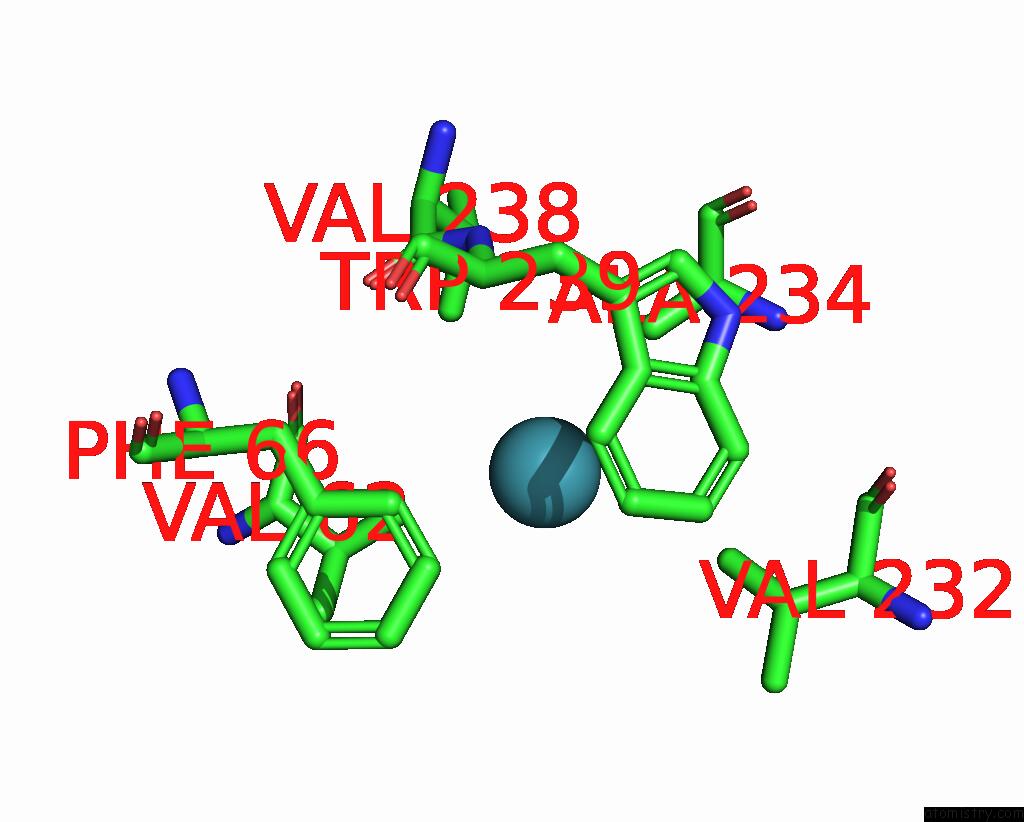
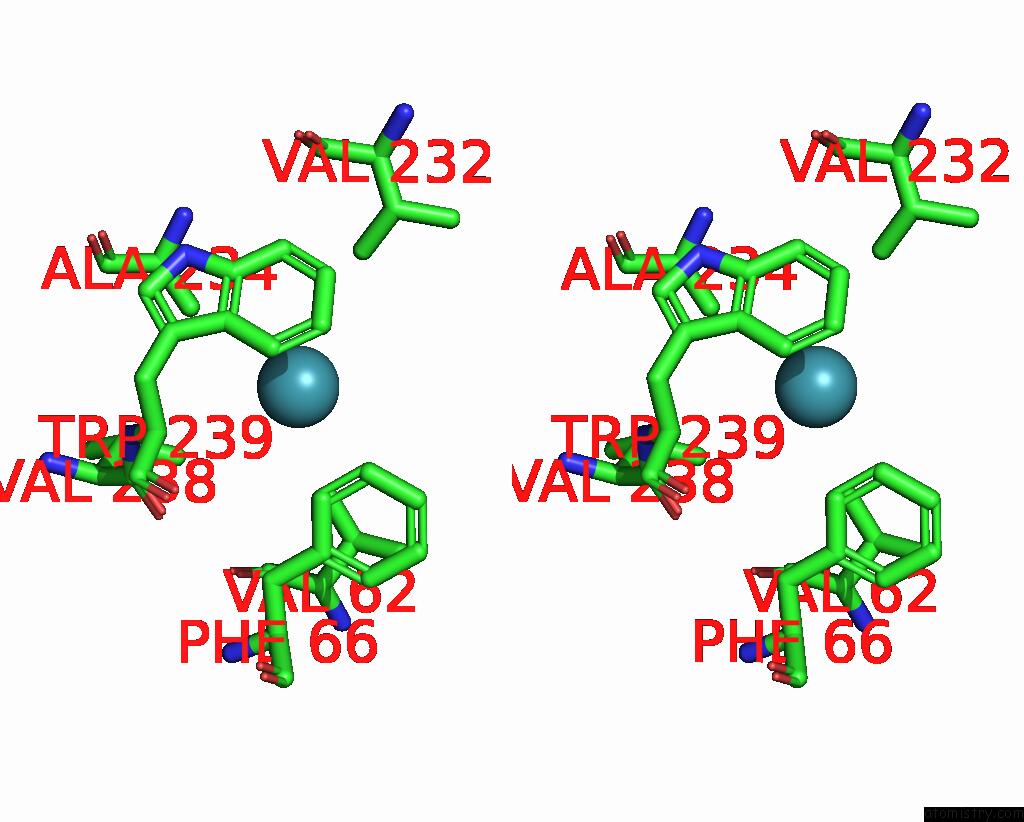
Xenon binding site 4 out of 5 in 7shw
Go back to
Xenon binding site 4 out
of 5 in the Crystal Structure of Mycobacterium Smegmatis Lmca with Xenon
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Xenon with other atoms in the Xe binding
site number 4 of Crystal Structure of Mycobacterium Smegmatis Lmca with Xenon within 5.0Å range:
|
Xenon binding site 5 out of 5 in 7shw
Go back to
Xenon binding site 5 out
of 5 in the Crystal Structure of Mycobacterium Smegmatis Lmca with Xenon
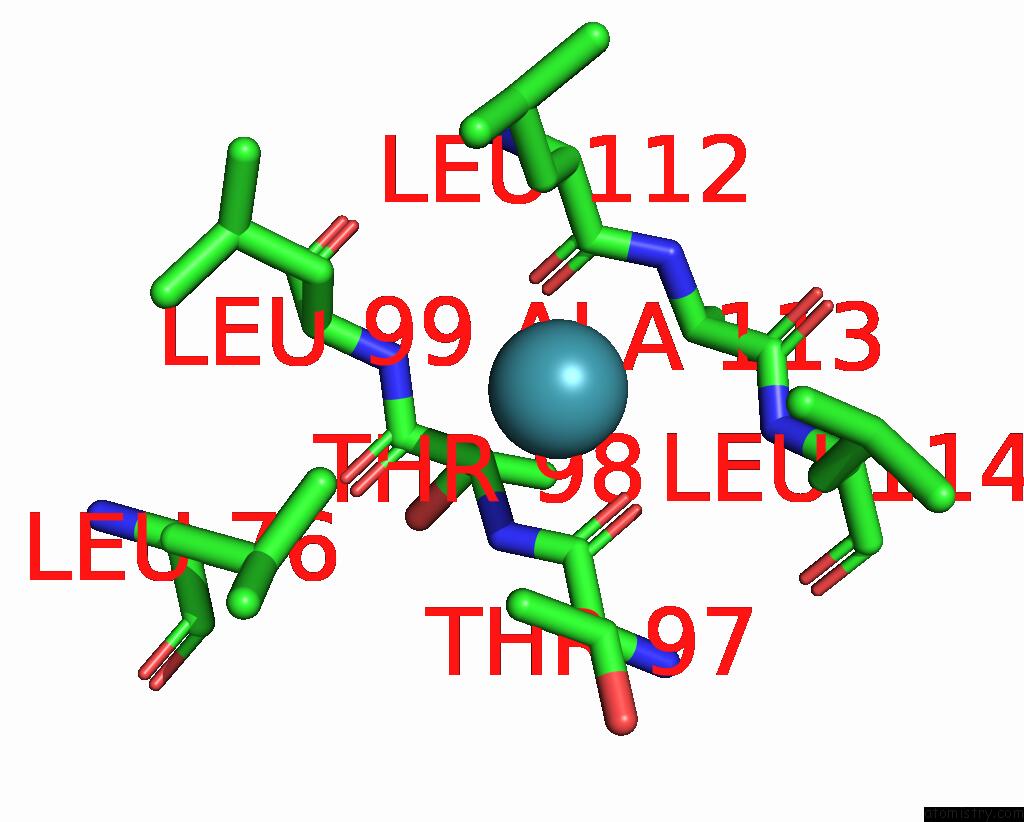
Mono view
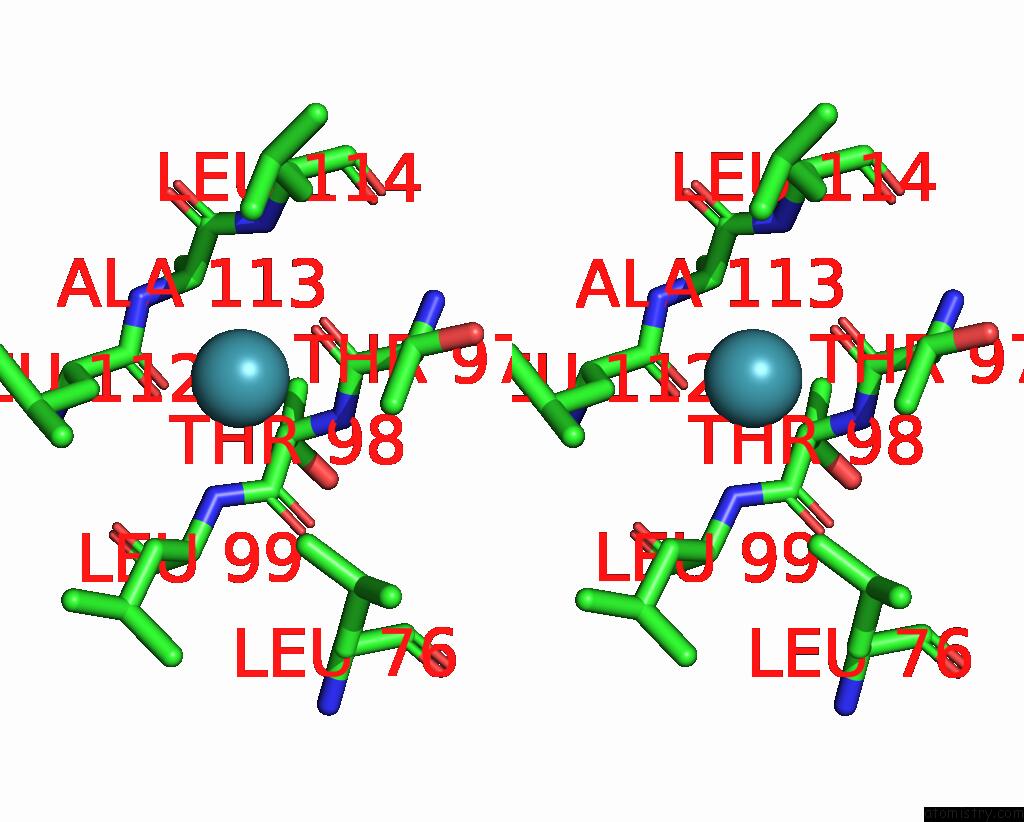
Stereo pair view

Mono view

Stereo pair view
A full contact list of Xenon with other atoms in the Xe binding
site number 5 of Crystal Structure of Mycobacterium Smegmatis Lmca with Xenon within 5.0Å range:
|
Reference:
O.Patel,
R.Brammananth,
W.Dai,
S.Panjikar,
R.L.Coppel,
I.S.Lucet,
P.K.Crellin.
Crystal Structure of the Putative Cell-Wall Lipoglycan Biosynthesis Protein Lmca From Mycobacterium Smegmatis. Acta Crystallogr D Struct V. 78 494 2022BIOL.
ISSN: ISSN 2059-7983
PubMed: 35362472
DOI: 10.1107/S2059798322001772
Page generated: Tue Aug 19 18:25:30 2025
ISSN: ISSN 2059-7983
PubMed: 35362472
DOI: 10.1107/S2059798322001772
Last articles
Zn in 1MKDZn in 1MG0
Zn in 1MKM
Zn in 1ML2
Zn in 1MEY
Zn in 1MGO
Zn in 1MH2
Zn in 1MFS
Zn in 1MFT
Zn in 1MC5